Digital twins of the ocean
Digital twins are virtual representations of the physical world. They advance our understanding of the past, the present, and the future of marine ecosystems.

More coming soon…
—
Origin and function of metaorganisms
Every living being interacts with microorganisms. The nature and outcome of their interaction can be studied in the laboratory.

Molecular basis and evolutionary dynamics of C. elegans-microbiota interactions: Research project within the CRC1182, Evolutionary Ecology and Genetics group (Prof. Dr. Hinrich Schulenburg)
All animals and plants host a multitude of microorganisms, such as viruses, bacteria, archaea, and fungi. They influence their host’s nutrition, development, immunity, and evolution. We hypothesized that, on a short time scale, hosts select microbes with beneficial functions (and vice versa) and, on longer time scales, those host-selected microbes are evolutionary advantageous.
Find our recent publication on the assembly of C. elegans‘ gut microbiome here.
—
Helminth immune modulation
Outcomes of infection are shaped by the host, the parasite, and their environment – but to what extent?

Epidemiological traits in helminth infections of sticklebacks: incorporating evolutionary and ecological perspectives in the study of helminth immune modulation in vertebrate hosts: Research project at the Max Planck Institute (MPI) for Evolutionary Biology, Parasitology Group (Dr. Martin Kalbe)
Epidemiological traits are shaped through effects of the host, the parasite and by their interaction (Figure 1 shows a theoretical illustration of this assumption). The relative contribution of each of the interaction partners may differ along the infection process and depends on the coevolutionary history. Different environments may select for different defence strategies among host populations, which results in immunological heterogeneity (Lazzaro et al., 2009; Graham, 2013). Here, we assessed virulence patterns of a vertebrate host-parasite association on a large geographic scale to test to what extent heterogeneous environments may cause immunological heterogeneity.
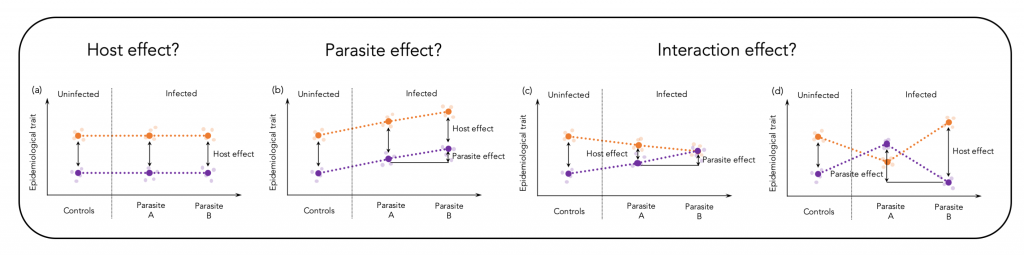

We hypothesized that different co-evolutionary trajectories of three-spined sticklebacks (Gasterosteus aculeatus) and their specific cestode parasite Schistocephalus solidus could be seen by specific virulence patterns on a geographic scale. Our data indeed provide evidence that differences in parasite prevalence can cause immunological heterogeneity and that parasite size, a proxy for virulence and resistance, is, on a geographic scale, determined by main effects of the host and the parasite and less by an interaction of both genotypes. The corresponding publications can be found here and here.

—
Phage-bacteria infection networks
Phages infect bacteria and can influence bacterial infection dynamics

Tripartite species interaction: infection dynamics of pipefish infecting vibrios and their temperate phages: Research project at the GEOMAR | Helmholtz Centre for Ocean Research Kiel, Pipefish group (Dr. Olivia Roth)
Bacteriophages (viruses that infect bacteria) play a significant role in host-parasite interactions: with up to 108 particles per mL in marine waters, viruses are the most abundant biological entity on earth and have supposedly profound effects on ecosystem composition and dynamics (Fuhrmann, 1999; Wilhelm and Suttle, 1999; Brockhurst et al., 2006; Vos at al., 2009; Stern and Sorek, 2011).
One type of bacteriophages are so-called temperate phages that have two different reproductive strategies. They either lyse the infected cell and disperse horizontally through burst of their host cell or they integrate into the bacterial chromosome and are vertically transmitted to the bacterial daughter cells (lysogenic cycle). Whether a temperate phage enters the lytic or the lysogenic cycle depends on environmental factors, e.g. temperature, nutrients, salinity, (UV) radiation and/or oxygen conditions (Kokjohn and Sayler, 1991; Jiang and Paul, 1994, 1996; Weinbauer et al., 2003).
We hypothesized a link between bacterial resistance to phages and bacterial harm to eukaryotic hosts (in this case pipefish Syngnathus typhle). We sampled Vibrio bacteria from pipefish, isolated vibriophages from the bacteria and constructed a replicated, fully reciprocal 75 × 75 phage-bacteria infection matrix. The phage-bacteria infection network showed a nested structure with some highly susceptible bacteria and some resistant bacteria, whereas most bacteria were susceptible to a small subset of phages (intermediate susceptible).
Our results further showed that phage-susceptible Vibrios were more harmful to the pipefish. We conclude that research across more than two species levels is needed for a thorough understanding of infection patterns and host-parasite evolution. The corresponding publication can be found here.
—
Chronic inflammatory diseases
Inflammation is a natural process to prevent infection. However, malfunctioning of the immune system can lead to chronic inflammation, allergies and disease

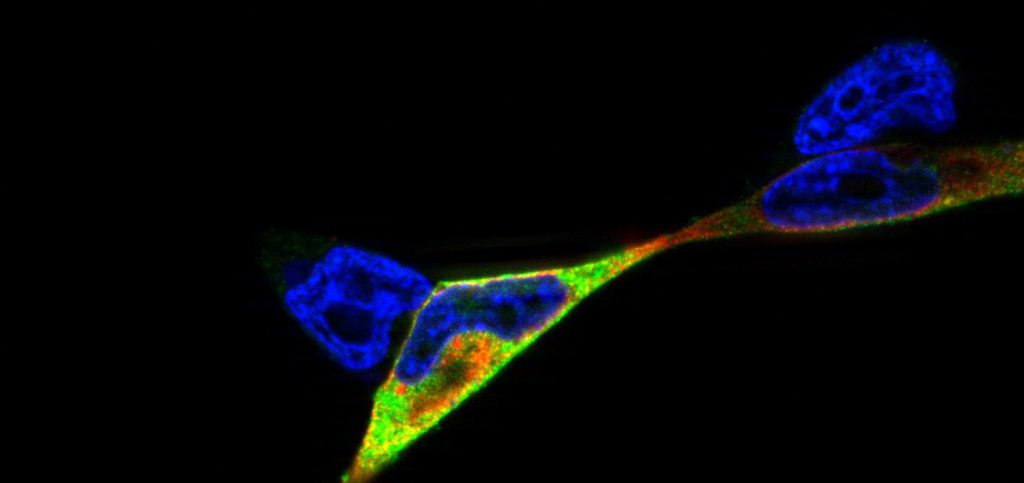
Studies of NOD2/ p22phox interaction: studying the interplay between innate immune receptors and ROS-producing enzymes in inflammatory disease: Research project at the Institute of Clinical Molecular Biology (icmb), cell biology group (supervision by Dr. Simone Lipinski, Prof. Dr. Philip Rosenstiel)
Reactive oxygen species (ROS) can be produced as a byproduct in biological reactions or as a primary function of the NADPH oxidase (NOX) complex. ROS-generating enzymes are evolutionary highly conserved and have been identified in a wide variety of organisms, including mammals, nematodes and plants (Kawahara et al., 2007). ROS play pivotal roles in cell signaling, host defence, and inflammation. Due to their high biochemical reactivity, ROS can protect against pathogens but they can also cause tissue damage by unspecific oxidation (oxidative stress).
Epithelial cells and mucosal surfaces form a direct interface between organism and invaders. We think that a crosstalk between ROS-producing enzymes and receptors of the innate immune system is critical to maintain the integrity and function of this part of the immune system. It has already been shown that inflammatory disorders such as the inflammatory bowel disease CD (Crohn’s disease) are connected with oxidative stress in affected tissues.
Here, we hypothesized that the innate immune receptor NOD2, which has been shown to play a role in the etiology of Crohn’s disease, interacts with the the NADPH oxidase component p22phox. We showed that NOD2 and p22phox co-localize upon activation and that there is a direct protein-protein interaction potentially involved in production of reactive oxygen species. The corresponding publication can be found here.